Discover how your daily coffee habit fuels gut health by fostering unique bacterial growth, unlocking new pathways to wellness!
Study: Coffee consumption is associated with intestinal Lawsonibacter asaccharolyticus abundance and prevalence across multiple cohorts. Image Credit: Shutterstock AI
Scientists identify a metabolic link between coffee consumption and the abundance of specific gut microorganisms across different human populations.
The study is published in the journal Nature Microbiology.
Background
Coffee is a popular beverage worldwide, with a range of health benefits. Studies have shown that coffee intake can reduce the risks of diabetes, non-alcoholic fatty liver disease, cancer, and all-cause and cardiovascular mortality.
The health benefits of coffee can be attributed to its polyphenol content. Chlorogenic acid, a key polyphenol in coffee, is metabolized by gut microorganisms into caffeic acid, quinic acid, and other metabolites. The gut microbiota that helps metabolize coffee has also been found to mediate its health effects. Several studies have reported that coffee intake can lead to changes in the gut microbiota composition and diversity in healthy individuals.
The scientists of the current study have previously reported that among more than 150 food items, coffee exhibits the highest correlation with the gut microbiota composition in over 1,000 individuals.
In the current study, scientists have conducted a multi-omic analysis of metagenomic samples obtained from more than 22,000 individuals who provided detailed reports on long-term coffee consumption. Additionally, they integrated these findings with public data from over 54,000 samples, encompassing diverse populations, including non-Westernized groups, newborns, and individuals with specific diseases.
Study Design
The study analyzed more than 35,000 metagenomic samples from three study cohorts, including the ZOE Personalized Responses to Dietary Composition Trial (PREDICT) metagenomics study, the Mind–Body Study (MBS), and the Men’s Lifestyle Validation Study (MLVS).
The study analysis also included more than 54,000 metagenomic samples from public sources, including healthy individuals, non-Westernized individuals, newborns and infants, ancient microbiome samples, non-human primates, and individuals with a specific disease.
Important Observations
The study investigated the association between coffee intake and gut microbiota composition by categorizing the participants into three coffee-drinking levels: never-drinking, moderate-drinking, and high-drinking.
The findings revealed a strong correlation between coffee intake and gut microbiota composition across different study populations. The gut microbiota exhibited distinct compositions in coffee drinkers compared to non-drinkers, with a modest effect on differentiating the level of coffee drinking.
In general, coffee showed stimulatory rather than inhibitory effects on the abundance of gut microbial species. The strongest association of coffee intake was observed with the abundance of the Gram-positive bacterium Lawsonibacter asaccharolyticus. This association remained the same for both decaffeinated and caffeinated coffee.
The abundance of Lawsonibacter asaccharolyticus was 4- to 8-fold higher among high coffee drinkers compared to that among non-drinkers. Among moderate drinkers, the abundance was 3- to 4-fold higher than non-drinkers.
The study conducted a set of in vitro experiments to confirm the observed association further. In these experiments, coffee was added to the culture media of Lawsonibacter asaccharolyticus.
The findings revealed that coffee significantly increased the growth of Lawsonibacter asaccharolyticus by 3.5-fold, irrespective of the types (moka brewed and instant coffee) and the presence of caffeine.
The study also identified a panel of 115 gut microbial species positively associated with coffee consumption, highlighting coffee’s broader influence on the microbiota.
Besides the coffee-mediated increased abundance of Lawsonibacter asaccharolyticus at the individual level, the study found that the overall prevalence of the bacterium in a population can be driven by the population-level consumption of coffee.
The analysis of 438 plasma metabolomes identified several metabolites enriched among coffee drinkers, with quinic acid and its potential derivatives associated with coffee and Lawsonibacter asaccharolyticus. Unannotated metabolites potentially derived from quinic acid were also highlighted, emphasizing the need for future biochemical investigations.
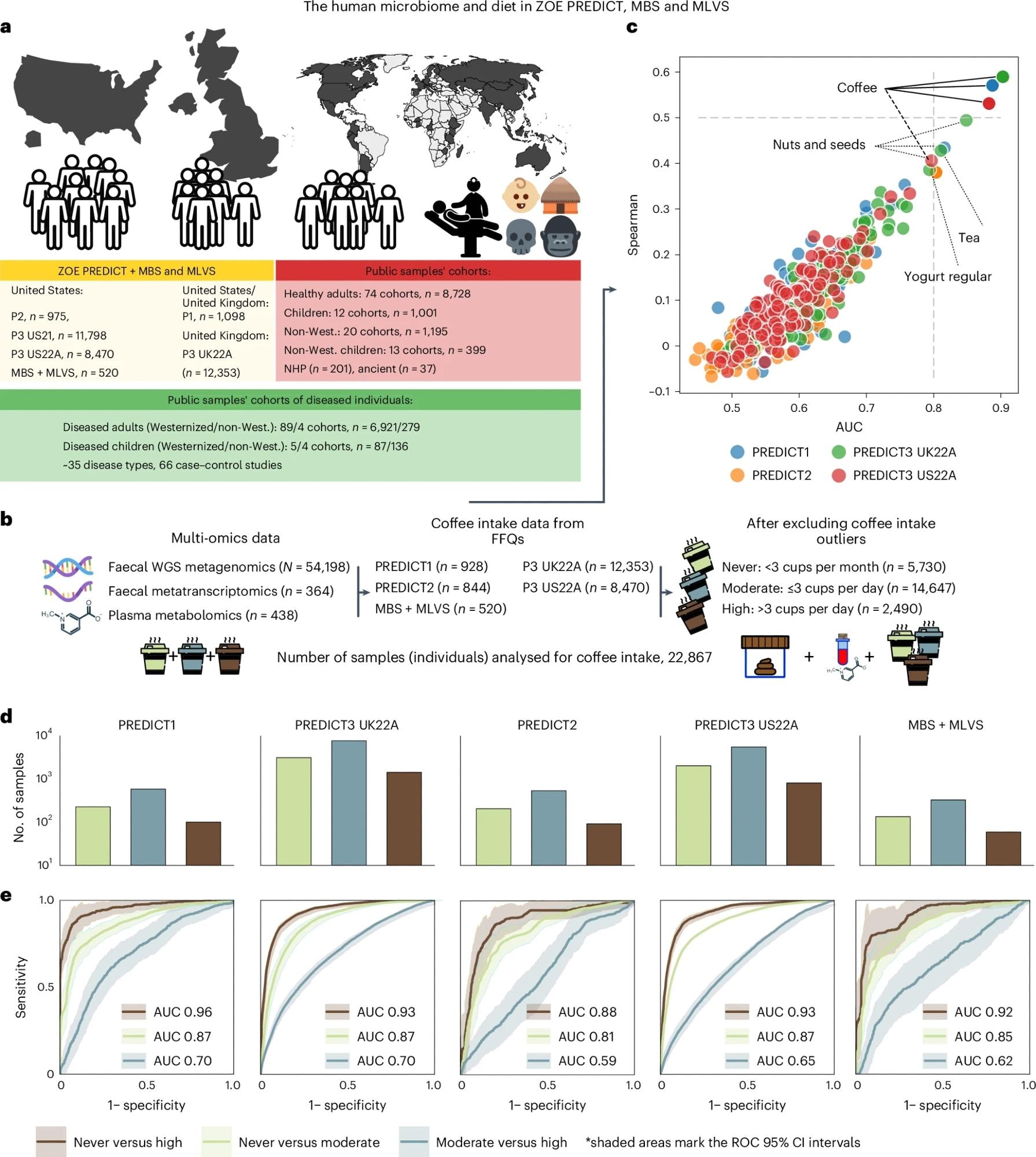 a, Five UK and/or US PREDICT cohorts (n = 975, 11,798, 8,470, 1,098, and 12,353), the MBS and the MLVS (n = 213 and n = 307, respectively) were used to assess diet–microbiome relationships (total n = 35,214). For later comparisons of microbiome distributions across different populations, we retrieved n = 18,984 metagenomic samples from public sources, including healthy adult individuals, newborns, non-Westernized (non-West.) individuals, ancient samples, and non-human primates (NHP). P1, PREDICT1; P2, PREDICT2; P3, PREDICT3. b, We combined fecal metagenomics (n = 54,198), fecal metatranscriptomics (n = 364), and plasma metabolomics (n = 438) with the latter two from the MBS and MLVS cohorts. FFQs surveyed the nutritional habits of the participants from four PREDICT cohorts, MBS, and MLVS (n = 22,867 after removing individuals above the 99th percentile of coffee intake in the PREDICT cohorts as outliers). Participants were categorized as ‘high’, ‘moderate’ and ‘never’ coffee drinkers as previously established25. c, Median Spearman’s correlation and median AUCs from a random forest regressor and a random forest classifier trained on the microbiome composition estimated by MetaPhlAn 4 . d, The number of never (light green), moderate (dark cyan) and high-coffee drinkers (brown). e, ROC and AUC of random forest classifiers discriminating participants between pairs of the three coffee drinker classes, assessed in a tenfold, ten times repeated cross-validations (CV) that benefited from the other cohorts during the training phase as in the leave-one-dataset-out approach (LODO). The shaded areas represent the 95% confidence intervals (CIs) of a linear interpolation over all the folds of the test.
a, Five UK and/or US PREDICT cohorts (n = 975, 11,798, 8,470, 1,098, and 12,353), the MBS and the MLVS (n = 213 and n = 307, respectively) were used to assess diet–microbiome relationships (total n = 35,214). For later comparisons of microbiome distributions across different populations, we retrieved n = 18,984 metagenomic samples from public sources, including healthy adult individuals, newborns, non-Westernized (non-West.) individuals, ancient samples, and non-human primates (NHP). P1, PREDICT1; P2, PREDICT2; P3, PREDICT3. b, We combined fecal metagenomics (n = 54,198), fecal metatranscriptomics (n = 364), and plasma metabolomics (n = 438) with the latter two from the MBS and MLVS cohorts. FFQs surveyed the nutritional habits of the participants from four PREDICT cohorts, MBS, and MLVS (n = 22,867 after removing individuals above the 99th percentile of coffee intake in the PREDICT cohorts as outliers). Participants were categorized as ‘high’, ‘moderate’ and ‘never’ coffee drinkers as previously established25. c, Median Spearman’s correlation and median AUCs from a random forest regressor and a random forest classifier trained on the microbiome composition estimated by MetaPhlAn 4 . d, The number of never (light green), moderate (dark cyan) and high-coffee drinkers (brown). e, ROC and AUC of random forest classifiers discriminating participants between pairs of the three coffee drinker classes, assessed in a tenfold, ten times repeated cross-validations (CV) that benefited from the other cohorts during the training phase as in the leave-one-dataset-out approach (LODO). The shaded areas represent the 95% confidence intervals (CIs) of a linear interpolation over all the folds of the test.
Study Significance
The study finds a strong correlation between coffee intake and gut microbiota composition across five US and UK populations.
A positive association has been observed between coffee intake and a set of 115 gut microbial species. Among these microorganisms, the strongest association has been observed for Lawsonibacter asaccharolyticus.
The observed stimulatory effect of coffee on the growth of Lawsonibacter asaccharolyticus provides a background for future studies aiming to decipher the extent of this stimulatory effect. Such studies should investigate the effect of different concentrations of coffee on the growth rates of a panel of coffee-associated gut microorganisms.
The study also identified variations in the prevalence of Lawsonibacter asaccharolyticus between Western and non-Western populations, potentially linked to differences in coffee availability and consumption habits.
The metabolomic analysis carried out in this study reveals that quinic acid, trigonelline, and other potential metabolites are significantly enriched in coffee drinkers carrying Lawsonibacter asaccharolyticus.
Chlorogenic acid is one of the main polyphenols in coffee. Gut microorganisms metabolize it to caffeic acid, quinic acid, and several other metabolites. Gut microorganisms responsible for this biotransformation include Bifidobacterium animalis, Bifidobacterium lactis, Escherichia coli, and Lactobacillus gasseri.
The caffeine-independent strong association between Lawsonibacter asaccharolyticus abundance and coffee intake indicates that it may also respond to activities within these polyphenol metabolism pathways.
Caffeine and its derivatives were prioritized in the study because of their association with Lawsonibacter asaccharolyticus. However, the enrichment of microorganisms in decaffeinated coffee drinkers indicates that caffeine does not occupy the central position in this complex crosstalk. Instead, quinic acid and its derivatives may play a pivotal role.
Overall, the study provides a framework for understanding microbial dietary responses at the biochemical level.
Journal reference:
- Manghi, P., Bhosle, A., Wang, K., Marconi, R., Ricci, L., Asnicar, F., Golzato, D., Ma, W., Hang, D., Thompson, K. N., Franzosa, E. A., Nabinejad, A., Tamburini, S., Rimm, E. B., Garrett, W. S., Sun, Q., Chan, A. T., Arumugam, M., Bermingham, K. M., . . . Song, M. (2024). Coffee consumption is associated with intestinal Lawsonibacter asaccharolyticus abundance and prevalence across multiple cohorts. Nature Microbiology, 9(12), 3120-3134. DOI: 10.1038/s41564-024-01858-9, https://www.nature.com/articles/s41564-024-01858-9