COMET, a novel machine learning framework, integrates EHR data and omics analyses using transfer learning, significantly enhancing predictive modeling and uncovering biological insights from small cohorts.
 Study: A machine learning approach to leveraging electronic health records for enhanced omics analysis. Image Credit: LeoWolfert / Shutterstock
Study: A machine learning approach to leveraging electronic health records for enhanced omics analysis. Image Credit: LeoWolfert / Shutterstock
In a recent study published in the journal Nature Machine Intelligence, researchers presented clinical and omics multimodal analysis enhanced with transfer learning (COMET), a deep learning and transfer learning protocol.
Omics technological advances have revolutionized biological understanding. Proteomic, metabolic, transcriptomic, and other assays have enabled cost-effective estimation of analytes in the same specimen. While these assays generate high-dimensional data, budgetary and clinical constraints limit the size of omics cohorts. Therefore, innovative approaches are needed to augment the analyses of high-dimensional data.
Although statistical methods address false positives, there are fewer methods for machine learning (ML). Some approaches rely on transfer learning, a technique wherein an ML model is learned from a pre-training dataset that is later used to study a smaller dataset. While more modern deep learning methods have been applied to statistical frameworks, they mainly rely on learning from informative metadata or omics data alone.
The COMET framework overcomes these limitations by integrating pretraining on large electronic health record (EHR) datasets and blending early and late fusion strategies, allowing improved predictive performance and biological discovery.
The study and findings
In the present study, researchers introduced COMET, a deep learning and transfer learning protocol that improves omics analyses. COMET could be applied when electronic health records (EHR) and omics data are available for a larger and smaller cohort. COMET comprises a method to embed longitudinal EHR data, pre-training, and multimodal modeling.
COMET involves an ML model trained solely on EHR data that will have its weights transferred to a multimodal architecture trained and evaluated on a smaller sample with omics and EHR data. First, COMET was applied to predict days to labor onset in a pregnancy cohort of over 30,904 individuals from Stanford Healthcare. Around 61 pregnant individuals (omics cohort) had multiple plasma samples throughout the last days of pregnancy, which were used to generate a proteomics dataset measuring 1,317 proteins.
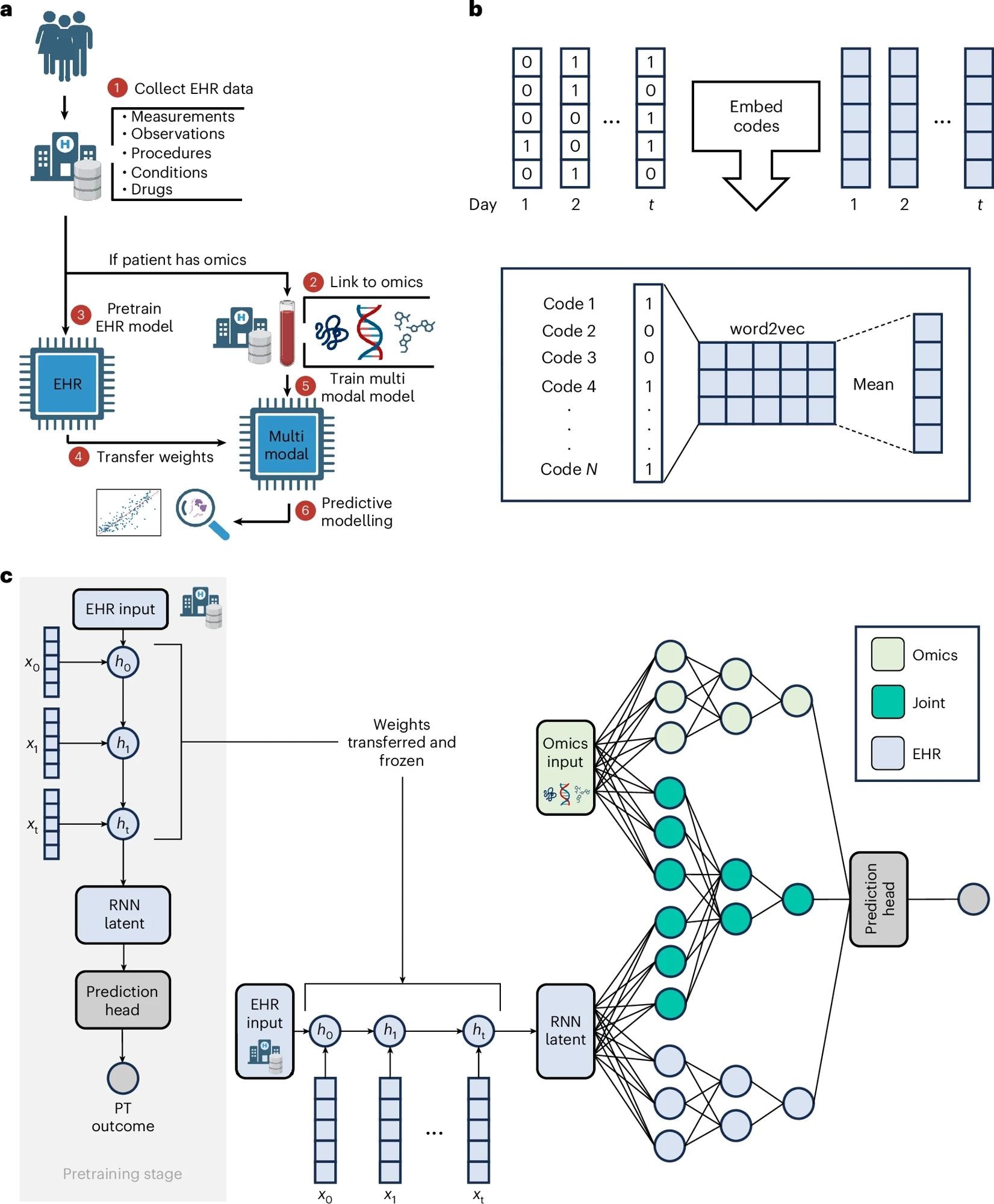 a, The input to COMET is EHR data and (for a subset of patients) paired, tabular omics data. The patients who only have EHR data are used to pretrain (PT) a neural network predict patient outcomes using only EHR data. The weights from this EHR network are transferred to a multimodal neural network used to analyse both EHR and omics data; the neural network is used for predictive modelling and post hoc analysis of the network is used for biological discovery. The COMET framework is flexible and can be used to predict any continuous or binary outcome. b, One-hot encoded vectors of EHR data (shown in white) are converted into embeddings (shown in blue) using word2vec; the embeddings for each code that occur within a particular day are averaged to compute sequential, summary embeddings. c, COMET uses a multimodal deep learning architecture to analyse both EHR data and omics data. Only EHR data are used in the pretraining stage; the core architecture is an RNN with gated recurrent units. After pretraining, the RNN weights are frozen and transferred into a multimodal architecture that analyses both EHR and omics data.
a, The input to COMET is EHR data and (for a subset of patients) paired, tabular omics data. The patients who only have EHR data are used to pretrain (PT) a neural network predict patient outcomes using only EHR data. The weights from this EHR network are transferred to a multimodal neural network used to analyse both EHR and omics data; the neural network is used for predictive modelling and post hoc analysis of the network is used for biological discovery. The COMET framework is flexible and can be used to predict any continuous or binary outcome. b, One-hot encoded vectors of EHR data (shown in white) are converted into embeddings (shown in blue) using word2vec; the embeddings for each code that occur within a particular day are averaged to compute sequential, summary embeddings. c, COMET uses a multimodal deep learning architecture to analyse both EHR data and omics data. Only EHR data are used in the pretraining stage; the core architecture is an RNN with gated recurrent units. After pretraining, the RNN weights are frozen and transferred into a multimodal architecture that analyses both EHR and omics data.
EHR data from the start of pregnancy through blood sampling were used to predict days to labor onset. After pre-training on EHR-only data (of 30,843 individuals), weights were transferred to a multimodal network trained to make predictions on the omics cohort. The model achieved a Pearson correlation coefficient of 0.868 (95% confidence interval [0.825, 0.900]), demonstrating its strong predictive capability. There was a strong correlation between the predicted days to labor onset and the actual number of days to labor onset, indicating that COMET was highly accurate in small cohorts with multidimensional data.
Next, COMET was compared with baseline models using only proteomics data, EHR data, or both. These baseline models solely used omics cohort data without pre-training. The EHR-only baseline model showed the worst performance, achieving a correlation of 0.768, while the proteomics-only model performed slightly better at 0.796. The joint baseline model was the best among the baselines, with a correlation of 0.815, albeit still inferior to COMET.
To gain deeper insights, researchers utilized t-distributed stochastic neighbor embedding (t-SNE) to visualize multimodal data by projecting the correlation matrix into two dimensions, revealing meaningful clusters of features based on their correlation patterns. Close features exhibit similar correlations with all other variables in the space. These clusters were annotated based on the medical concepts the EHR or protein features represent within each cluster. Various proteins showed significant correlations with EHR variables.
The team computed the feature importance for each protein. Proteins identified as highly significant in COMET models correlated with fetal development, pregnancy complications, and gestational age, aligning with established biological knowledge. Next, COMET was applied to a cancer cohort from the United Kingdom (UK) Biobank to predict three-year cancer mortality. Participants were all patients diagnosed with any cancer within five years of enrollment.
A subset of participants had blood samples available that were analyzed for proteomics data. They were included in the omics cohort if the samples were collected within one year of cancer diagnosis. Consistently, COMET achieved superior performance in predicting three-year cancer mortality compared to all baselines, with an area under the receiver operating characteristic curve (AUROC) of 0.842, significantly outperforming the joint baseline (AUROC 0.786) and single-modality models. The prevalence of three-year mortality in the omics cohort was 5.5%.
Further, t-SNE was used to visualize the correlation matrix, which revealed less overlap between EHR and proteomics data modalities in contrast to labor onset data. Nevertheless, there were significant correlations between EHR and proteomics data modalities when the correlation network was visualized, with each modality individually projected into two dimensions. Mortality factor 4-like protein 2 exhibited the strongest correlations with EHR features, particularly drug prescriptions, highlighting its potential as a prognostic biomarker.
A vast proportion of cancer patients’ proteins (66%) showed no correlation with any EHR variable. Further, the researchers estimated the correlation between each EHR feature and all proteins and the maximum correlation across all proteins for each EHR feature. This revealed many EHR features with low correlations to proteins in cancer patients, underscoring the value of including several data modalities.
Proteins with greater feature importance in COMET models aligned with known cancer prognostic biomarkers. Importantly, nine proteins that were more significant in COMET models were statistically associated with mortality status, further validating the model’s biological relevance.
Conclusions
In sum, the study illustrated the ability of COMET to augment predictive modeling across multiple tasks through pre-training and transfer learning. COMET yielded better-regularized models, which more accurately reflected known biology. Moreover, COMET models identified biologically relevant proteins for specific health outcomes.
In labor onset models, COMET revealed proteins crucial for pregnancy complications, immune regulation, and placental development, with Pearson correlation values supporting its predictive strength. For cancer mortality, identified proteins were those involved in tumor proliferation and microenvironment modulation. Overall, COMET provides a foundation for delineating complex relationships between clinical phenotypes and molecular mechanisms.
Journal reference:
- Mataraso SJ, Espinosa CA, Seong D, et al. A machine learning approach to leveraging electronic health records for enhanced omics analysis. Nature Machine Intelligence, 2025, DOI: 10.1038/s42256-024-00974-9, https://www.nature.com/articles/s42256-024-00974-9